Explore the latest insights from top science journals in the Muser Press daily roundup (August 17, 2025), featuring impactful research on climate change challenges.
In brief:
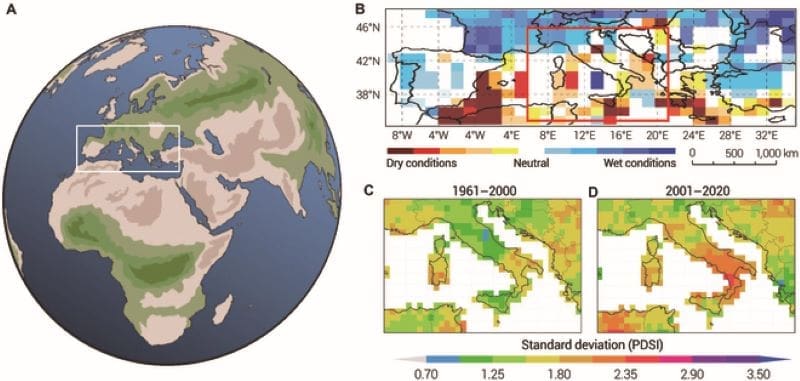
Mediterranean climate’s future: a swing between droughts and downpours
From its olive groves to its coastal cities, the Mediterranean depends on a delicate balance of rain and sun, but climate change is tipping the scales.
Using a new and relatively simple data-driven model, a study published in Ocean-Land-Atmosphere Research forecasts repeating drought and rain cycles in the central Mediterranean, offering a sharper picture of the region’s shifting water patterns. The findings pave the way for smarter, more adaptive strategies for facing growing climate volatility.
Long known for its sunny skies and temperate climate, the Mediterranean is now on the frontlines of climate change. The region is experiencing more frequent and intense climatic extremes, from droughts to downpours, prompting scientists to ramp up efforts to forecast future shifts in this culturally rich, agriculturally critical zone.
“While conventional Global Circulation Models have greatly advanced our understanding of climate dynamics, they don’t always capture key regional details, especially temperature variations that influence local rainfall patterns,” said Gianni Bellocchi, senior scientist at Université Clermont Auvergne in France and corresponding author of this study.
The team therefore aimed for a rather local lens into wetting and drying cycles in the central Mediterranean.
“We integrated climate forcings with historical data using a data-driven model with exogenous data, or DDMX,” said Nazzareno Diodato, first author and fellow scientist at the Met European Research Observatory – International Affiliates’ Program of the University Corporation for Atmospheric Research.
The researchers analyzed more than a century’s worth of historical weather data, dating back to 1884, with a focus on the Standardized Precipitation Index, a key indicator that tracks unusual wet or dry conditions over six-month intervals. Their DDMX model combines large-scale climate drivers with regional sea-level pressure data to improve predictive accuracy.
When validated against observations from 1993 to 2022, the model showed strong performance, with a correlation coefficient of 0.58 and a root mean square error of 0.621, confirming its reliability.
“Our DDMX approach offers a simpler methodology and complementary alternative to conventional Global Circulation Models for understanding and forecasting climate-related phenomena,” said Rajib Maity, co-author and researcher at the Indian Institute of Technology Kharagpur.
The model projects a decade-long drying period from 2045 to 2054, followed by a wetter phase – capturing the alternating wet–dry cycles that are emerging as a dominant signature of climate change in the region. These findings assist with water resource planning, agriculture, and infrastructure investment.
The trio plans to further refine and expand the DDMX model across different Mediterranean sub-regions and hydroclimate scenarios. They also aim to integrate more diverse datasets, explore new machine learning architectures, and quantify the uncertainty in forecasts more rigorously.
“Ultimately, we hope that our work will lead to more effective resilience planning and sustainable water management strategies in regions such as the Mediterranean that are particularly vulnerable to evolving climate patterns,” Bellocchi said.
Journal Reference:
Nazzareno Diodato, Rajib Maity, Gianni Bellocchi, ‘Persistent Oscillations in the Decadal Prediction of Central Mediterranean Wetting and Drying Phases’, Ocean-Land-Atmosphere Research 4: 0093 (2025). DOI: 10.34133/olar.0093
Article Source:
Press Release/Material by Ocean-Land-Atmosphere Research (OLAR)
Scientists hack microbes to identify environmental sources of methane
Roughly two-thirds of all emissions of atmospheric methane – a highly potent greenhouse gas that is warming planet Earth – come from microbes that live in oxygen-free environments like wetlands, rice fields, landfills and the guts of cows.
Tracking atmospheric methane to its specific sources and quantifying their importance remains a challenge, however. Scientists are pretty good at tracing the sources of the main greenhouse gas, carbon dioxide, to focus on mitigating these emissions. But to trace methane’s origins, scientists often have to measure the isotopic composition of methane’s component atoms, carbon and hydrogen, to use as a fingerprint of various environmental sources.
A new paper by researchers at the University of California, Berkeley, reveals how the activity of one of the main microbial enzymes involved in producing methane affects this isotope composition. The finding could change how scientists calculate the contributions of different environmental sources to Earth’s total methane budget.
“When we integrate all the sources and sinks of carbon dioxide into the atmosphere, we kind of get the number that we’re expecting from direct measurement in the atmosphere. But for methane, large uncertainties in fluxes exist – within tens of percents for some of the fluxes – that challenge our ability to precisely quantify the relative importance and changes in time of the sources,” said UC Berkeley postdoctoral fellow Jonathan Gropp, who is first author of the paper. “To quantify the actual sources of methane, you need to really understand the isotopic processes that are used to constrain these fluxes.”
Gropp teamed up with a molecular biologist and a geochemist at UC Berkeley to, for the first time, employ CRISPR to manipulate the activity of this key enzyme to reveal how these methanogens interact with their food supply to produce methane.
“It is well understood that methane levels are rising, but there is a lot of disagreement on the underlying cause,” said co-author Dipti Nayak, UC Berkeley assistant professor of molecular and cell biology. “This study is the first time the disciplines of molecular biology and isotope biogeochemistry have been fused to provide better constraints on how the biology of methanogens controls the isotopic composition of methane.”
Many elements have heavier or lighter versions, called isotopes, that are found in small proportions in nature. Humans are about 99% carbon-12 and 1% carbon-13, which is slightly heavier because it has an extra neutron in its nucleus. The hydrogen in water is 99.985% hydrogen-1 and 0.015% deuterium or hydrogen-2, which is twice as heavy because it has a neutron in its nucleus.
The natural abundances of isotopes are reflected in all biologically produced molecules and variations can be used to study and fingerprint various biological metabolisms.
“Over the last 70 years, people have shown that methane produced by different organisms and other processes can have distinctive isotopic fingerprints,” said geochemist and co-author Daniel Stolper, UC Berkeley associate professor of earth and planetary science. “Natural gas from oil deposits often looks one way. Methane made by the methanogens within cow guts looks another way. Methane made in deep sea sediments by microorganisms has a different fingerprint.
“Methanogens can consume or ‘eat’, if you will, a variety of compounds including methanol, acetate or hydrogen; make methane; and generate energy from the process. Scientists have commonly assumed that the isotopic fingerprint depends on what the organisms are eating, which often varies from environment to environment, creating our ability to link isotopes to methane origins.”
“I think what’s unique about the paper is, we learned that the isotopic composition of microbial methane isn’t just based on what methanogens eat,” Nayak said. “What you ‘eat’ matters, of course, but the amount of these substrates and the environmental conditions matter too, and perhaps more importantly, how microbes react to those changes.”
“Microbes respond to the environment by manipulating their gene expression, and then the isotopic compositions change as well,” Gropp said. “This should cause us to think more carefully when we analyze data from the environment.”
The paper was published in the journal Science.
Vinegar- and alcohol-eating microbes
Methanogens – microorganisms that are archaea, which are on an entirely separate branch of the tree of life from bacteria – are essential to ridding the world of dead and decaying matter. They ingest simple molecules – molecular hydrogen, acetate or methanol, for example – excreted by other organisms and produce methane gas as waste.
This natural methane can be observed in the pale Will-o’-the-wisps seen around swamps and marshes at night, but it’s also released invisibly in cow burps, bubbles up from rice paddies and natural wetlands and leaks out of landfills. While most of the methane in the natural gas we burn formed in association with hydrocarbon generation, some deposits were originally produced by methanogens eating buried organic matter.
The isotopic fingerprint of methane produced by methanogens growing on different “food” sources has been well established in laboratory studies, but scientists have found that in the complexity of the real world, methanogens don’t always produce methane with the same isotopic fingerprint as seen in the lab. For example, when grown in the lab, species of methanogens that eat acetate (essentially vinegar), methanol (the simplest alcohol), or molecular hydrogen (H2) produce methane, CH4, with a ratio of hydrogen and carbon isotopes different from the ratios observed in the environment.
Gropp had earlier created a computer model of the metabolic network in methanogens to understand better how the isotope composition of methane is determined. When he got a fellowship to come to UC Berkeley, Stolper and Nayak proposed that he experimentally test his model. Stolper’s laboratory specializes in measuring isotope compositions to explore Earth’s history. Nayak studies methanogens and, as a postdoctoral fellow, found a way to use CRISPR gene editing in methanogens. Her group recently altered the expression of the key enzyme in methanogens that produces the methane – methyl-coenzyme M reductase (MCR) – so that its activity can be dialed down. Enzymes are proteins that catalyze chemical reactions.
Experimenting with these CRISPR-edited microbes – in a common methanogen called Methanosarcina acetivorans growing on acetate and methanol – the researchers looked at how the isotopic composition of methane changed when the enzyme activity was reduced, mimicking what is thought to happen when the microbes are starved for their preferred food.
They found that when MCR is at low concentrations, cells respond by altering the activity of many other enzymes in the cell, causing their inputs and outputs to accumulate and the rate of methane generation to slow so much that enzymes begin running both backwards and forwards.
In reverse, these other enzymes remove a hydrogen from carbon atoms; running forward, they add a hydrogen. Together with MCR, they ultimately produce methane (CH4). Each forward and reverse cycle requires one of these enzymes to pull a hydrogen off of the carbon and add a new one ultimately sourced from water. As a result, the isotopic composition of methane’s four hydrogen molecules gradually comes to reflect that of the water, and not just their food source, which starts with three hydrogens.
This is different from typical assumptions for growth on acetate and methanol that assume no exchange between hydrogen derived from water and that from the food source.
“This isotope exchange we found changes the fingerprint of methane generated by acetate and methanol consuming methanogens vs. that typically assumed. Given this, it might be that we have underestimated the contribution of the acetate-consuming microbes, and they might be even more dominant than we have thought,” Gropp said. “We’re proposing that we at least should consider the cellular response of methanogens to their environment when studying isotopic composition of methane.”
Beyond this study, the CRISPR technique for tuning production of enzymes in methanogens could be used to manipulate and study isotope effects in other enzyme networks broadly, which could help researchers answer questions about geobiology and the Earth’s environment today and in the past.
“This opens up a pathway where modern molecular biology is married with isotope-geochemistry to answer environmental problems,” Stolper said. “There are an enormous number of isotopic systems associated with biology and biochemistry that are studied in the environment; I hope we can start looking at them in the way molecular biologists now are looking at these problems in people and other organisms – by controlling gene expression and looking at how the stable isotopes respond.”
For Nayak, the experiments are also a big step in discovering how to alter methanogens to derail production of methane and redirect their energy to producing useful products instead of an environmentally destructive gas.
“By reducing the amount of this enzyme that makes methane and by putting in alternate pathways that the cell can use, we can essentially give them another release valve, if you will, to put those electrons, which they were otherwise putting in carbon to make methane, into something else that would be more useful,” she said.
Other co-authors of the paper are Markus Bill of Lawrence Berkeley National Laboratory and former UC Berkeley postdoc Rebekah Stein, and Max Lloyd, who is a professor at Penn State University. Gropp was supported by a fellowship from the European Molecular Biology Organization. Nayak and Stolper were funded, in part, by Alfred B. Sloan Research Fellowships. Nayak also is an investigator with the Chan-Zuckerberg Biohub.
Journal Reference:
Jonathan Gropp et al., ‘Modulation of methyl–coenzyme M reductase expression alters the isotopic composition of microbial methane’, Science 389, 6761, pp. 711-715 (2025). DOI: 10.1126/science.adu2098
Article Source:
Press Release/Material by Robert Sanders | University of California – Berkeley (UC Berkeley)
Climate models reveal human influence behind stalled Pacific cycle
The recent North Pacific Ocean temperatures oceanic shifts are directly linked to the prolonged megadrought gripping the American Southwest, and this research published in Nature suggests it may not ease for another 30 years.
“Our results show that the drought and ocean patterns we’re seeing today are not just natural fluctuations – they’re largely driven by human activity,” said Jeremy Klavans, postdoctoral researcher in CU Boulder’s Department of Atmospheric and Oceanic Sciences and lead author of the study.
Cracking a long-standing climate puzzle
For over a century, scientists have tracked a climate cycle in the North Pacific known as the Pacific Decadal Oscillation (PDO), which alternates every few decades between a warm “positive” phase and a cool “negative” phase. In its negative phase – where cooler waters hug the U.S. West Coast – storm tracks shift northward and rainfall in the western U.S. decreases significantly.
Until now, the PDO was assumed to be governed almost entirely by natural internal processes, such as air-sea interactions. Even the most recent Intergovernmental Panel on Climate Change (IPCC) report stated with high confidence that the PDO is not influenced by human activity.
But using a new suite of over 500 climate model simulations, Klavans and his team found that since the 1950s, over half of the variability in the PDO can be attributed to human emissions, including both greenhouse gases and aerosols. Prior to 1950, these patterns were mostly driven by natural processes.
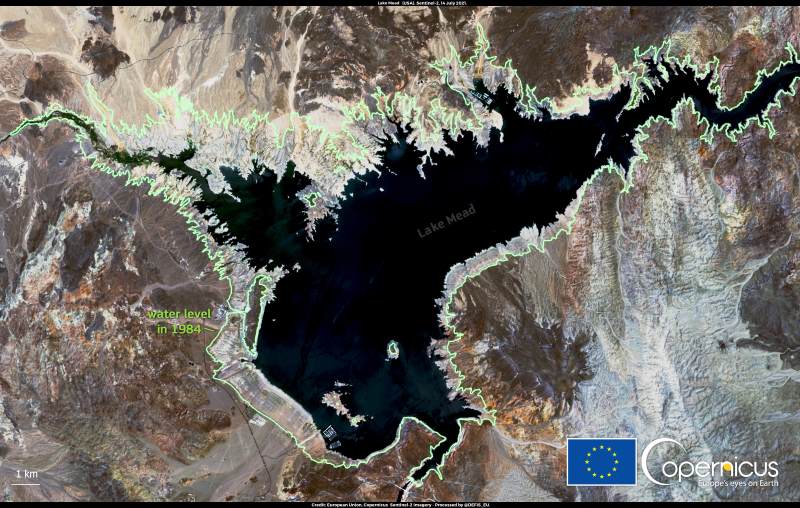
The PDO is stuck – and that’s a problem
The PDO has remained locked in a negative phase since the 1990s – an unusually long period for what is typically a fluctuating cycle. This prolonged cool phase has been a major driver of the ongoing megadrought in the western U.S., drying out the region by pushing precipitation-bearing storms farther north and reducing overall moisture in the air.
“If the PDO were purely natural, we would have expected it to shift back to positive after the strong El Niño in 2015,” Klavans said. “Instead, it flipped briefly and then reverted – suggesting something deeper and undiscovered is holding it in place.”
New tools, new understanding
This breakthrough was made possible by advances in climate modeling that helped scientists correct for an issue in models known as the “signal-to-noise paradox.” This study finds that most climate models have historically overestimated natural variability while underestimating the effects of human-driven external forcing.
“Once we corrected for that imbalance, it became clear that human emissions are the dominant factor behind the current PDO pattern and the West’s extreme dryness,” Klavans said.
Pedro DiNezio, a professor at the University of Colorado and co-author of the study, added: “Our work shows that on decadal timescales, climate models have been underestimating how sensitive regional climates are to external forcing.”
Worst drought in over 1,200 years – and no relief in sight
The implications of this work are sobering. Research shows the American Southwest is in the midst of its driest 20-year period in at least 1,200 years. About 93% of the western U.S. is currently in drought, with 70% experiencing severe conditions.
The study warns that if greenhouse gas emissions continue at current rates, the PDO will likely stay in its negative phase, and the region’s water crisis will deepen.
“This isn’t a temporary dry spell,” Klavans said. “It’s a climate-driven transformation of the region’s water system. Planners and policymakers need to treat it as such.”
Global consequences
The findings may also extend beyond the Pacific. Similar patterns exist in other ocean basins – like the North Atlantic Oscillation (NAO), which is tied to droughts in parts of Europe, including Spain.
“Our methods have the potential to drastically improve predictions of climate impacts – including precipitation trends across the globe,” added Amy Clement, a professor at the University of Miami and co-author of this study. “That kind of foresight is critical for planning and adapting to a changing climate.”
Journal Reference:
Klavans, J.M., DiNezio, P.N., ‘Clement, A.C. et al., ‘Human emissions drive recent trends in North Pacific climate variations’, Nature (2025). DOI: 10.1038/s41586-025-09368-2
Article Source:
Press Release/Material by University of Colorado at Boulder (CU Boulder)
New toolbox for breeding climate-resilient crop plants
An international research team headed by Heinrich Heine University Düsseldorf (HHU) and the Max Planck Institute for Plant Breeding Research in Cologne (MPIPZ) has developed a new and very precise method for identifying so-called genomic regulatory switches. These switches are responsible for the manifestation of plant traits.
In the scientific journal Nature Genetics, the researchers describe that, although these regulatory switches make up only a small fraction of the genome, they can have a significant influence on plant traits. The team demonstrated the method on regulatory switches relating to drought stress, identifying promising starting points for the breeding of new maize varieties adapted to e.g. climate change.

Natural genetic variation in the genome ensures biodiversity and drives evolution. However, as natural evolutionary processes require millennia, we cannot wait for them to adapt crop plants to the rapidly changing climatic conditions, which are responsible for e.g. increased drought periods. To safeguard global food security, researchers must accelerate the identification of appropriate natural DNA variants to improve crop plant performance under stress conditions.
A research team headed by Dr Thomas Hartwig and Dr Julia Engelhorn from the Institute for Molecular Physiology at HHU and MPIPZ now presents a new, efficient method for mapping the genetic “switches” of plants. Not actually genes themselves, these small sections of the genome determine when, where and to what extent a gene is active. They are comparable with a dimmer switch regulating the brightness of a lamp.
While research so far largely focused on the genes themselves, the new study demonstrates that key differences between plants – e.g. variation in size, or resistance to diseases or stress situations – are often not determined by the genes, but rather by these regulatory switches. Traditionally, however, it is not only difficult to locate these regions precisely, but also to determine which changes play the decisive role. This is now changing thanks to a new, scalable mapping method developed within the framework of the project.
The research team analysed 25 different maize hybrids, i.e. crossbreeds of different maize varieties, identifying over 200,000 regions in the genome where natural variations influence regulatory switches.
Dr Julia Engelhorn, lead author of the study: “Although these regulatory switches make up less than 1% of the genome, the variations often explain a substantial share of heritable trait differences – sometimes exceeding half.”
Dr Thomas Hartwig, corresponding author of the study, comments: “Understanding how these regulatory switches operate provides a powerful new tool to enhance both crop resilience and yield – laying the foundation for smarter breeding processes in the future.”
The researchers applied their method specifically to traits, which play a role in drought stress, identifying over 3,500 individual regulatory switches and the associated genes via which the plants respond to water-limited conditions.
Engelhorn: “Our approach allows direct comparison of the differences in switch variants inherited via the maternal and paternal lines in a single experiment. We can thus offer the maize research community a resource of over 3,500 drought-linked regulatory sites – opening up new possibilities to fine-tune gene expression for enhanced robustness.”
Hartwig: “The precision of this mapping enables us to learn from the natural differences in the switches how they work, which in turn enables targeted manipulation of the switches to develop plants with improved traits.”
This research was realised in collaboration with a team from the University of California in Davis, in which Dr Samantha Snodgrass is a member. The co-author of the study emphasises the change in perspective accompanying the approach: “Despite decades of successful research, much of the genome – the parts outside the genes – remains a black box. This new method pulls back the curtain and enables us to identify the function of these non-coding areas, providing biologists and breeders with new, precise targets for new research and development approaches.”
***
The study was conducted within the CEPLAS Cluster of Excellence on Plant Sciences at HHU and MPIPZ. Other sources of funding include the European Horizon Europe project BOOSTER, which aims to advance the development of climate-resilient cereal crops.
Journal Reference:
Engelhorn, J., Snodgrass, S.J., Kok, A., Seetharam, A.S., Schneider, M., Kiwit, T., Singh, A., Banf, M., Khaipho-Burch, M., Runcie, D.E., Camargo, V.S., Torres-Rodriguez, J.V., Sun, G., Stam, M., Fiorani, F., Schnable, J.C., Bass, H.W., Hufford, M.B., Stich, B., Frommer, W.B., Ross-Ibarra, J., Hartwig, T., ‘Genetic variation at transcription factor binding sites largely explains phenotypic heritability in maize’, Nature Genetics (2025). DOI: 10.1038/s41588-025-02246-7
Article Source:
Press Release/Material by Heinrich-Heine University Duesseldorf (HHU)
Featured image credit: Gerd Altmann | Pixabay